Scientists uncover nuclear droplets that link multiple leukemias revealing new therapeutic target
A hidden structure inside the cell is rewriting how scientists understand leukemia. Beneath the microscope, what looked like disorder turned out to follow a simple physical rule – one that connects several major mutations behind the disease.
In new research from şŁ˝ÇÉçÇř, scientists have discovered that different genetic drivers of leukemia use the same secret compartments inside the cell nucleus to keep cancer growing. The finding points to a shared physical target that could inspire new kinds of treatments.
The work reshapes a long-standing view of how a common leukemia begins and offers a fresh way to design therapies that strike a single weakness shared across distinct genetic forms of the disease.
Leukemia starts when mutations in blood-forming cells disrupt the balance between growth and differentiation. Patients with entirely different genetic changes show strikingly similar patterns of gene activity and can respond to the same drugs.
What invisible thread could make so many mutations behave the same way?
To find out, the Riback and Goodell labs at Baylor joined forces. Dr. Joshua Riback, an assistant professor and who studies how proteins form droplets through a process known as phase separation, teamed with Dr. Margaret “Peggy” Goodell, Baylor’s chair of the Department of Molecular and Cellular Biology and a pioneer in understanding how blood stem cells give rise to leukemia. Together, they set out to follow the physics hidden inside cancer’s chemistry.
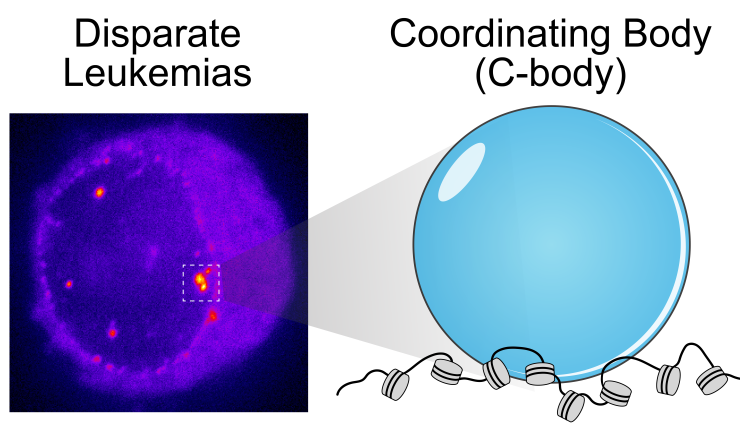
Then came the moment of clarity. Graduate student Gandhar Datar, co-mentored by Riback and Goodell, peered into Riback’s high-resolution microscope and saw something no one expected: leukemia cell nuclei shimmered with a dozen bright dots – tiny beacons missing from healthy cells.
Those dots weren’t random. They contained large amounts of mutant leukemia proteins and drew in many normal cell proteins to coordinate activation of the leukemia program. The dots were new nuclear compartments formed by phase separation, the same physical principle that describes why oil droplets form in water. The team named this new compartment, “coordinating bodies,” or C-bodies.
Inside the nucleus, these C-bodies act like miniature control rooms, pulling together the molecules that keep leukemia genes switched on. Like drops of oil collecting on the surface of soup, they appear when the cell’s molecular ingredients reach just the right balance.
Even more surprising, cells carrying entirely different leukemia mutations formed droplets with the same behavior. Although their chemistry differs, the resulting nuclear condensates perform the same function, using the same physical playbook.
A new quantitative assay developed in the Riback lab confirmed it. These droplets are biophysically indistinguishable – like soups made from different ingredients that still simmer into the same consistency. No matter which mutation started the process, each leukemia formed the same kind of C-body.
“It was astonishing,” Riback said. “All these different leukemia drivers, each with its own recipe, ended up cooking the same droplet, or condensate. That’s what unites these leukemias and gives us a common target. If we understand the biophysics of the C-body, its general recipe, we’ll know how to dissolve it and reveal new insights for targeting many leukemias.”
The team confirmed the finding across human cell lines, mouse models and patient samples. When they tweaked the proteins so they could no longer form these droplets – or dissolved them with drugs, the leukemia cells stopped dividing and began to mature into healthy blood cells.
“Seeing C-bodies in patient samples made the link crystal clear,” said co-author Elmira Khabusheva, a postdoctoral associate in the Goodell lab. “By putting existing drugs into the context of the C-body, we can see why they work across different leukemias and start designing new ones that target the condensate itself. It’s like finally seeing the whole forest instead of just the trees.”
“By identifying a shared nuclear structure that all these mutations depend on, we connect basic biophysics to clinical leukemia,” added Goodell. “It means we can target the structure itself – a new way of thinking about therapy.”
“Across every model we studied, the pattern was the same,” Datar said. “Once we saw those bright dots, we knew we were looking at something fundamental.”
The discovery of C-bodies gives leukemia a physical address a structure scientists can now see, touch and target. It provides a simple physical explanation for how different mutations converge on the same disease and points to treatments aimed at dissolving the droplets that cancer depends on – like skimming the fat from a soup to restore its balance.
This finding sets up a new paradigm for linking droplet-forming disease drivers into shared, generalizable therapeutic targets, revealing that just as distinct mutations in leukemia converge on the same condensate, other diseases, such as ALS, may each assemble their own biophysically indistinguishable droplets governed by the same physical rules.
The discovery was made possible through collaboration between the Riback and Goodell labs at şŁ˝ÇÉçÇř and international partners including the Associazione Italiana per la Ricerca sul Cancro (AIRC), the Trond Mohn Foundation and the Norwegian Cancer Society.
The study, led by Gandhar Datar and Elmira Khabusheva, appears in the journal .
Others who contributed to the research include: Archish Anand, Joshua Beale, Marwa Sadek, Chun-Wei Chen, Evdokiia Potolitsyna, Nayara Alcantara-Contessoto, Guangyuan Liu, Josephine De La Fuente, Christina Dollinger, Anna Guzman, Alejandra Martell, Katharina Wohlan, Abhishek Maiti, Nicholas J. Short, S. Stephen Yi, Vibeke Andresen, Bjørn Tore Gjertsen, Brunangelo Falini, Rachel E. Rau, Lorenzo Brunetti, and Nidhi Sahni. See for affiliations.
This project was supported by CA183252 (M.A.G.), CA26574 (M.A.G.) and F30CA268725 (G.K.D.). J.A.R. is a CPRIT Scholar in Cancer Research with funding from the Cancer Prevention and Research Institute of Texas (CPRIT) New Investigator Grant RR210040 and was supported by The Ted Nash Long Life Foundation, the Leukemia Research Foundation, and the Searle Scholars Program. R.E.R. was supported by K08CA201611 and K12CA090433-11. B.F. was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC). L.B. was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC) Start-up Grant nr.22895. VA was supported by the Trond Mohn Foundation and The Research Council of Norway (project no. 303358). BTG was supported by the Norwegian Cancer Society (grants numbers 303445 and 19017), The Research Council of Norway (project no. 303358) and Helse Vest Health Trust (project no. F-13102). See for additional funding information.



 Credit
Credit